 Home
Home
 Back
Back
Definition: This calculator computes the concentration of a nucleic acid sample (DNA or RNA) based on its absorbance at 260 nm (\( A_{260} \)), using the Beer-Lambert Law.
Purpose: It is used in molecular biology to determine the concentration of DNA or RNA samples prior to experiments like PCR, cloning, or sequencing, ensuring optimal performance in downstream applications.
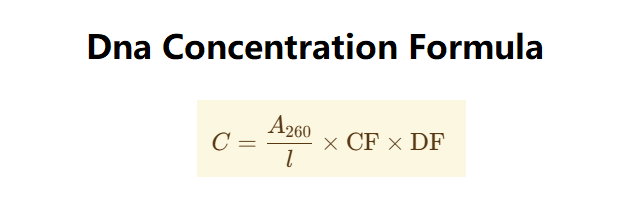
The calculator uses the following formula:
\( C = A_{260} \times \text{conversion factor} \times \text{dilution factor} \)
Where:
Steps:
Calculating the concentration of nucleic acids is crucial for:
Example 1: Calculate the concentration of a dsDNA sample with an \( A_{260} \) of 2.5, undiluted:
Example 2: Calculate the concentration of an ssRNA sample with an \( A_{260} \) of 1.2, diluted 10-fold, outputting in ng/µL:
Q: Why is absorbance measured at 260 nm?
A: Nucleic acids like DNA and RNA absorb UV light most strongly at 260 nm due to their conjugated bases, making it the standard wavelength for concentration measurements.
Q: What if my sample is contaminated with RNA or proteins?
A: This calculator does not account for contaminants. RNA and proteins can also absorb at 260 nm, leading to overestimation. Use absorbance ratios like \( A_{260}/A_{280} \) (around 1.8 for pure DNA, 2.0 for pure RNA) to assess purity separately.
Q: Can this calculator be used for oligonucleotides?
A: Yes, but the conversion factors (33 µg/mL for ssDNA, 40 µg/mL for ssRNA) are approximate for oligonucleotides. For more accuracy, calculate the extinction coefficient based on the specific sequence.