 Home
Home
 Back
Back
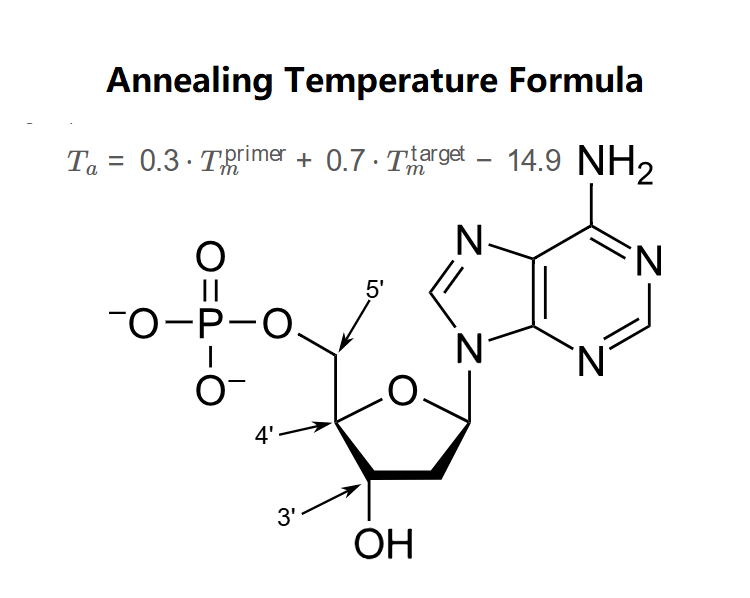
Definition: This calculator computes the optimal annealing temperature (\( T_a \)) for the annealing step in a Polymerase Chain Reaction (PCR) based on the melting temperatures of the primer (\( T_m^{\text{primer}} \)) and the target DNA (\( T_m^{\text{target}} \)).
Purpose: It is used in molecular biology to ensure specific and efficient primer binding to the target DNA during PCR, maximizing amplification yield and specificity.
The calculator uses the following empirical formula:
\( T_a = 0.3 \cdot T_m^{\text{primer}} + 0.7 \cdot T_m^{\text{target}} - 14.9 \)
Where:
Steps:
Calculating the optimal annealing temperature is crucial for:
Example 1: Calculate the annealing temperature for a primer with \( T_m^{\text{primer}} = 60 \, \text{°C} \) and target DNA with \( T_m^{\text{target}} = 80 \, \text{°C} \):
Example 2: Calculate the annealing temperature for a primer with \( T_m^{\text{primer}} = 333.15 \, \text{K} \) (60°C) and target DNA with \( T_m^{\text{target}} = 353.15 \, \text{K} \) (80°C), outputting in Kelvin:
Q: Why is the annealing temperature critical in PCR?
A: The annealing temperature determines the specificity of primer binding. Too low, and primers may bind to unintended sequences, causing nonspecific amplification. Too high, and primers may not bind efficiently, reducing yield.
Q: How do I determine the melting temperatures (\( T_m \))?
A: Calculate \( T_m \) based on primer sequence, GC content, and salt concentration using standard bioinformatics tools or software provided by primer suppliers.
Q: Can I use this calculator for non-PCR applications?
A: Yes, but the formula is optimized for PCR. For other techniques like Southern blotting, you may need to adjust the annealing temperature based on experimental conditions.